Working with Helper Phages
Choosing a helper phage
What is a helper phage?
Plasmids carrying the intergenic region of filamentous phage (oriF1) can package as ssDNA in viral particles in the presence of a phage (1). When wild-type phages are used, interference of the plasmid with the phage replication leads to reduction in the phage copy number and drastic decrease in virion production (2). Helper phages are designed to overcome interference, maximize virion production and keep packaging of their own ssDNA at a low level.
M13KO7
M13KO7 is a derivative of M13 containing an origin of replication P15A and a kanamycin resistance gene inserted in the domain B of the origin of replication (3). The decrease in phage production resulting from the disruption of domain B is compensated by the P15A origin and a Met40Ile mutation in gene II (reported in (3) but not found in our isolate). The kanamycin resistance gene allows for selection of cells superinfected by the helper. M13KO7 is a remarkable helper phage with a plasmid versus phage packaging of 10 to 1 (Fig. 1) and high-level production of plasmid-containing particles for DNA sequencing, mutagenesis (Kunkel-based methods) and phage display.
Other Helpers
R408 is a derivative of f1 that does not have any antibiotic selection marker and was initially reported to improve packaging of ssDNA (4). VCSM13 is a derivative of M13KO7 originally commercialized by Stratagene and often used in phage display; the phenotype of VCSM13 has never been published and exact sequence is unclear. All other published helper phages (e.g. hyperphage, R408d3, KM13, etc.) are derivatives of M13KO7, VCSM13 or R408.
CM13
We analyzed the phenotype of different known interference-resistance mutations in M13KO7 background, including irA1, a G->A mutation at position 8250 located 20 nucleotides before the ATG codon of gene II, and irB1, a C->T mutation at position 143 causing a Thr->Ile change in gene II protein (2). We also considered two mutations reported in VCSM13 gene II sequence; a G->T mutation located at the same position than irA1, and a A->G mutation at position 8419 causing an Ile->Val change in gene II protein; by analogy, we named these mutations irA3 and irB3. We could not confirm the presence of ir3A in our isolate of VCSM13. Both M13KO7/irA1+ and M13KO7/irA3+ mutants have highly increased virion production, but cells tend to stop growing, making the control of superinfection very difficult. M13KO7/irB1+and M13KO7/irA1+irB1+both failed to produce significant amount of virions. M13KO7/irB3+ gave a phenotype identical to VCSM13, producing often more virions than M13KO7 and giving a large-plaque phenotype on TG1. The amount of rf DNA (dsDNA replicative form) per cells is higher and phagemids are packaged at a 3:1 ratio (Fig. 1). M13KO7/irB3+is sold under the name CM13 on our catalog.
CM13 versus VCM13
VCSM13 as also a large plaque phenotype, a higher rf DNA content and a lower packaging ratio (Fig. 1). The M13KO7/irAB3+ double mutant appears similar to the single mutant M13KO7/irB3+ (Fig. 1) and, in this context, the mutation irB3 seems to be solely responsible for the helper phenotype. In practice, CM13 and VCSM13 are indistinguishable.
CM13 or M13KO7
CM13 gives larger plaques and yields twice more cfu/ml in production; it is therefore easier to prepare. CM13 consistently gives good phage yields, even at low multiplicity of infection, but has a lower packaging ratio. M13KO7 produces similar phage yields, requires bacteria to be precisely around 0.5 OD600 for superinfection and gives a high packaging ratio. Both M13KO7 & CM13 helpers can be used for phage display. M13KO7 is the recommended choice for preparing ssDNA, e.g. for Kunkel mutagenesis, because of its better packaging ratio
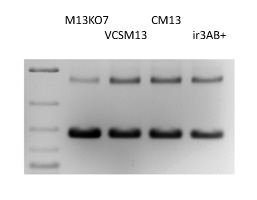
Figure 1. Helper Phage Packaging. Circular ssDNA from pADL-10b/scFv virions produced by different helper phages were analyzed by gel electrophoresis; Size indicated by the dsDNA ladder on the left lane are irrelevant.
Protocol for Transduction using M13KO7 or CM13
Conditions for optimal transduction
Always use bacteria freshly made; typically grow your phagemid-containing bacteria overnight in 2xYT medium from a single colony in the presence of ampicillin and glucose at 30°C or 37°C. Dilute the bacterial culture 1:20 v/v with fresh 2xYT medium and incubate for one hour at 37°C. Measure the absorbance at 600 nm, at best using a large 1 ml cuvette and a 1:5 or a 1:10 dilution; adjust to 0.5 OD and add the helper between 1 x 10(9) and 1 x 10(10) pfu/ml. We recommend 2 x 10(9) pfu/ml or 1 µl of our concentrated helper preparations per ml of culture as a good balance between not using too much helper and achieving a high level of superinfection; more helper may yield more phage but results are often inconsistent.
Immediately transfer the culture to a shaker and incubate for 30 min to 1 h at 37°C and 250 rpm; preincubation of helper and bacteria on the bench or at 37°C without shaking is unnecessary and often leads to a greater variability in phage yields; best superinfections are obtained at 37°C and 250 rpm. Then add kanamycin 50 µM, ampicillin 100 µM and IPTG 200 µM if you are using pADL-10b, pAK100 or pAK200 phagemid and lower the temperature to 30°C; harvest the phage after 8 h to o/n.
Virion production, cell density and immunity to superinfection
We have determined that the optimal cell culture density to add the helper is between 0.4 and 0.5 OD600 for TG1 and SS320 cells. Higher densities may leave a large percentage of non-transduced bacteria not producing virions while lower densities may amplify disparities caused by differences in phage growth rates and lower phage production as well when expression of toxic fusion proteins limits bacterial growth. It is known that phage infection and expression of the N-terminal domains of protein III inhibit superinfection (5). Because expression of protein III fusions is not well-controlled is many phagemids, the use of a truncated protein III lacking N-terminal domains has become a popular means to overcome immunity to superinfection. Practically, we have not seen a difference in virion production between full-length protein III phagemids (e.g. pHEN2 or pADL-23c) and truncated protein III phagemids (e.g. pCOMB3) when superinfection is conducted at the recommended cell density.
References
-
Dotto GF, Enea V, Zinder ND (1981). Functional analysis of bacteriophage f1 intergenic region. Virol 114:463-473.
-
Enea, V. and Zinder, N. (1982) Interference resistant mutants of phage f1, Virology 122, pp. 222-226.
-
Vieira, J. and Messing, J. (1987) R. Wu and L. Grossman (Eds.), Methods Enzymol., 153, pp. 3-11. San Diego: Academic Press.
-
Russel, M., Kidd, S., & Kelley, M. R. (1986). An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene, 45(3), 333–8.
-
Boeke, J.D., P. Model, and N.D. Zinder. (1982). Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol. Gen. Genet. 186:185-192.
PUB006 – Rev140404
Please send comments to info@abdesignlabs.com.
Copyright © 2014 Antibody Design Labs. All rights reserved. The reuse or reproduction of any of the information, design or layout contained in this web site without the permission of Antibody Design Labs is prohibited.
